Shoghi Lab
Projects
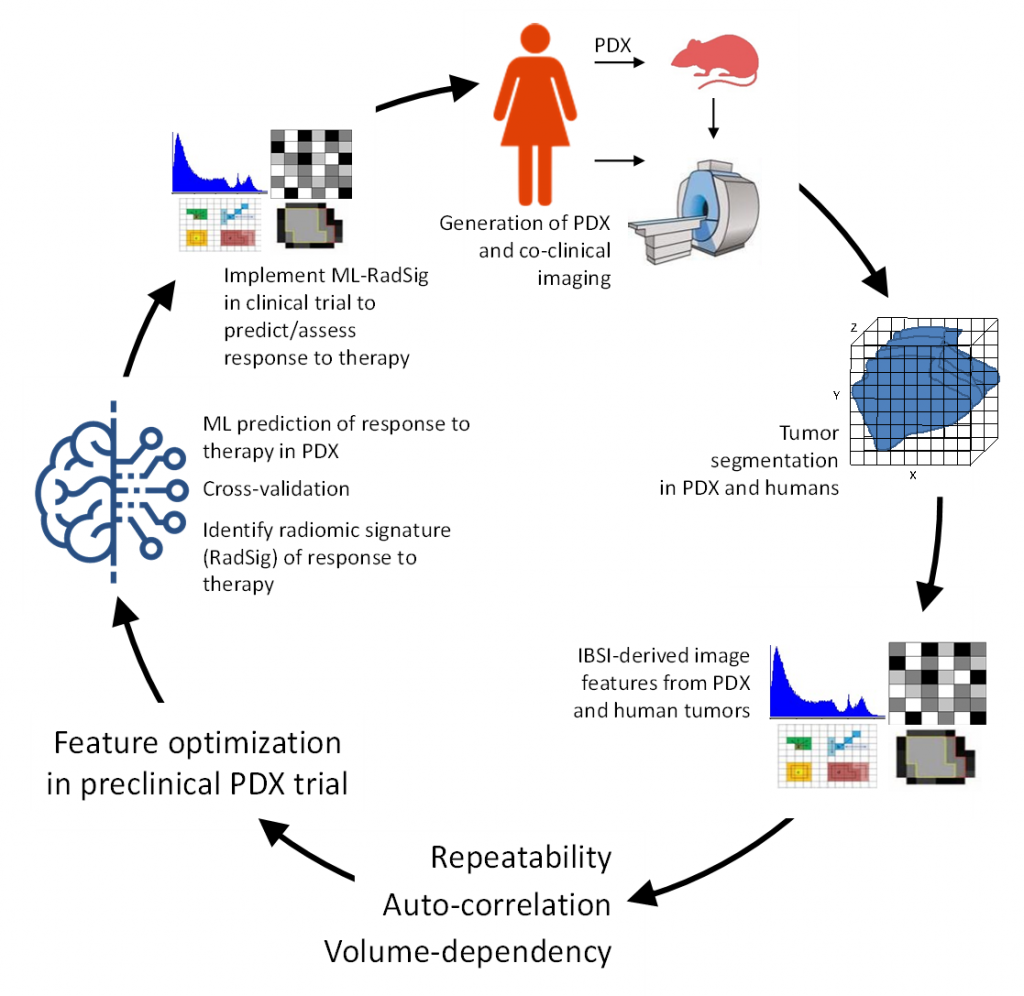
Co-Clinical Imaging Research
Multidisciplinary effort to facilitate the promise of precision medicine by optimizing the use imaging and realistic animal models of cancer (e.g., PDX, GEMM) to better inform of therapeutic outcome in clinical trials. The overall goal of the Co-Clinical Imaging Research Resource (C2IR2) is to develop and implement advanced quantitative imaging (QI)/radiomic pipelines to predict response to therapy in TNBC, and integrate to QI/radiomic image features with multi-scale analytic data (-OMICS, path) via machine learning algorithms to enhance prediction. For more information, visit the C2IR2 website.
In our most recent renewal application, we are developing quantitative imaging techniques to:
- Predict response to therapy in advanced ER+ breast cancer.
- Targeted PARP1 imaging and therapy in DNA damage response pathways.
- Imaging of metabolism and inflammation/oxidative stress axis in cancer pathogenesis and impact on therapeutic outcome.
Computational Imaging & Biology
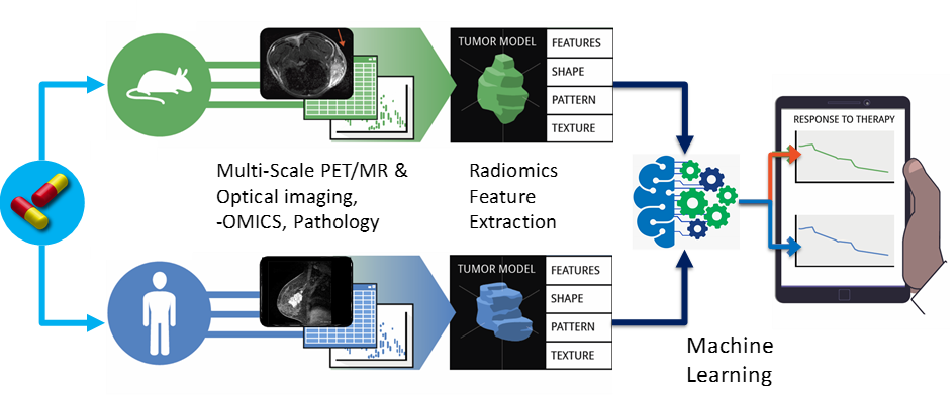
The Shoghi Lab is developing technology to capture the heterogeneity of diseased tissue via radiomic analyses and computational imaging methods in both preclinical and clinical research. These methods are primarily investigated in the context of co-clinical imaging research. Current projects include:
- Artificial intelligence and machine learning techniques to characterize imaging and extract biological insights.
- Multiscale imaging relating imaging features derived in vivo imaging, ex vivo imaging, multi-omics, and multiplexed pathology imaging to integrate imaging/biology across scales.
- Developing computational biology models (aka in silico biology or digital twins) to integrate with quantitative imaging. The purpose of the models is to define therapeutic endpoints.
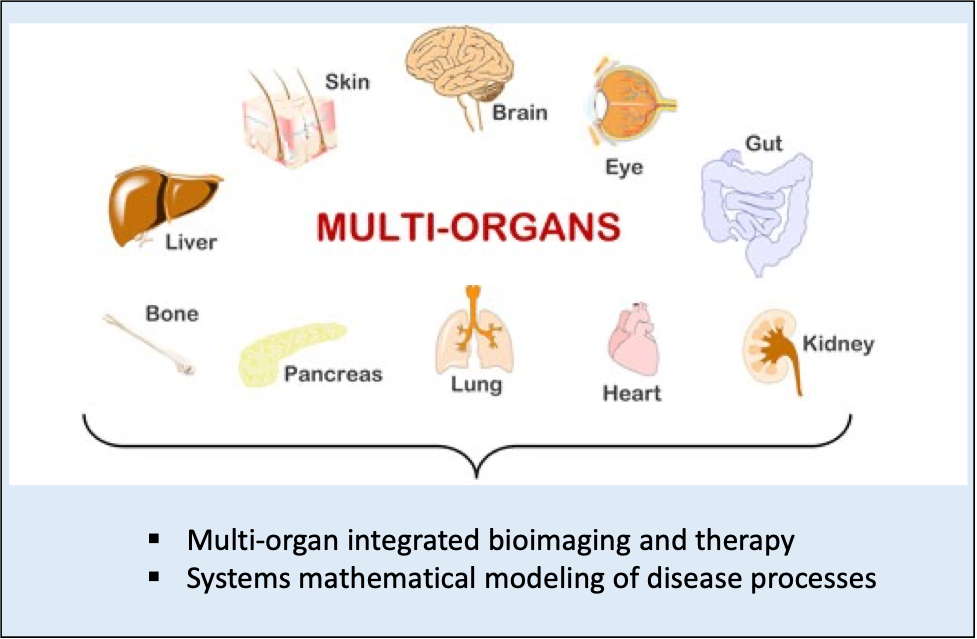
Multi-Systems Quantitative Imaging & Modeling
Obesity and Type-2-Diabetes (T2D) are a result of systemic disturbances in metabolism and inflammation. In light of the highly interconnected and coordinated nature of substrate metabolism in health, and its failure in disease, the goal of the project is to elucidate imaging biomakers for the progression of obesity/diabetes using preclinical animal models of diabetes with a translational endpoint. Of particular interest, is application of imaging metabolism/inflammation at the interface with obesity/diabetes and neurological diseases.
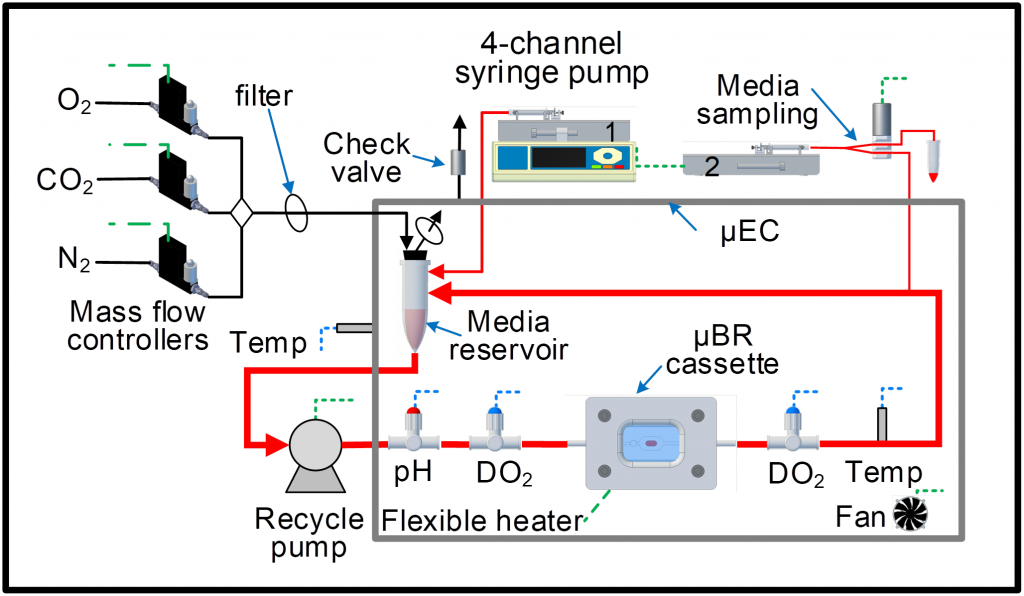
Ex-Vivo Artificial Tissue Bioreactor
An artificial tissue bioreactor is a versatile system designed to simulate the 3-dimensional (3D) structure and microenvironment of tissues in vivo. In vivo, the responses of individual cells are regulated by spatiotemporal cues that reside in the local microenvironment such as the extracellular matrix (ECM), neighboring cells, soluble factors and physical forces, all presented in a 3D context. When cells are isolated from their in-vivo environment, they are usually placed in a monolayer environment with limited cell-cell contact and bathed in a static medium, thus spatiotemporal cues are lost resulting in a multitude of cells displaying phenotypic instability. We are currently developing a mobile artificial tissue bioreactor to be integrated with imaging instruments to facilitate research, discovery, and validation of therapeutic and imaging biomarkers.
Preclinical Imaging Informatics
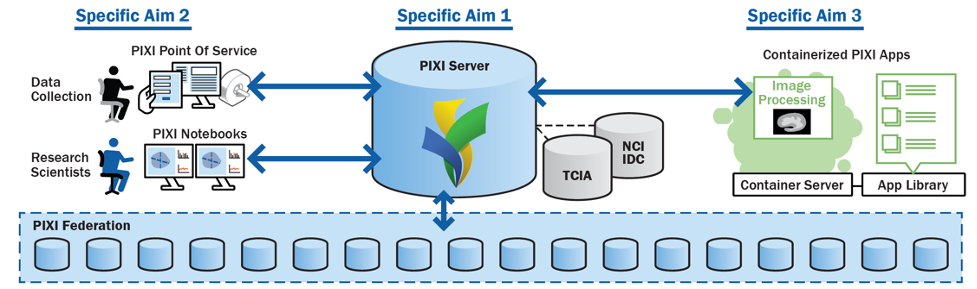
In collaboration with the Neuroinformatics Research Group (NRG), led by Daniel Marcus, PhD, we are developing an open source preclinical imaging XNAT-enabled informatics (PIXI) platform to manage the workflow of preclinical imaging and to develop computational imaging pipelines in the cloud. Please see the PIXI website updates on progress and opportunities to become an early adopter, and eventually download the open-source platform.